AU:
Compare single/double mutants from literature and prioritize purification and analysis based on this.
Add Ceftazidime in the growth assay
Is there any correlation between MIC and activity? may be ratio of WT-MIC with Mutant and WT-activity with Mutant?
See in the literature if there is any difference between His-Tag and Non-His tag b-lactamase or His-tag was removed before assay.
Forgot to mention: As Ping is back, I will start looking into extension data to make graph.
Self Note: Take everything to group meeting
AU-Group Meeting 11-11-2014.ppt
XG:
*Draft a response letter to Enzo to address (1) inpurity issue (2) proper labeling on SDS-PAGE gel picture (3) quote for the cost of enzyme with high purity ..
*PLOS One data analysis for different NAD+ concentrations
*Get quote for SIRT1, 2, and 3 plasmid from Origene
*Literature work
(1) maximum inhibition by NAM for all these sirtuins
(2) All known info on rate constants of all these sirtuins in the same table including which step is rate limiting if known
(3) Also make a table with all known mutants and activity effects, including which mutants were studied computationally
(4) NAM inhibition study on Sir2Af2
PMC-AT Group Meeting_11.11.2014.pptx
RC:
-Please update the tables indicated above as the become available (perhaps indicate which dropbox folder they can be found in)
-Also, plan an isoNAM/NAM series of initial rate experiments as discussed (analogous to DHP/NAM). Here you should choose a single isoNAM
concentration where we see activation in presence of NAM. This [isoNAM] will be fixed while [NAM] is varied.
Let me know when the planning is complete.
XG(11.14):
*A table for all known SIRTUIN mutants are listed in excel file below. It needs a lot of time to search and generate.
SIRTUIN_Mutants.xlsx
RC: Most of these mutants are characterized in terms of biological activity; however, we are interested in enzyme mechanism in vitro. You can omit the former in the future. Should we conclude that there is very little mutagenesis data for enzyme kinetics in vitro for enzymes other than Sir2Tm?
XG(11/20: Yes. Among the sirtuin mutagenesis work,very little part of them are focusing enzyme kinetics.
RC: For example, some of the QM/MM studies referenced mutagenesis experiments on Sir2. Did you get all of those? (As noted earlier we are ultimately looking for the cross-referencing of whatever experimental work and computational work there is on the roles of various residues. Ping was asked to comment on some of the latter.)
Also, I believe that Steegborn, as noted, has some papers (including one in 2013) where he looked at mammalian SIRT mutants.
There could be others.
XG(11/20): I do not think file "SIRTUIN_Mutants" has covered all of the studies. It depends on how you search, such as keyword-based, or author/research group oriented...I used "Web of Science" database through UPenn library system, then key in "sirtuin", "mutant"... to find the list. Other way to search is to list all the SIRTUIN research group PI's name, then key in "specific sirtuin's name" and "PI name", then search. I can do another search based on the second way.
RC: See below regarding revisiting of C pocket binding affinity/Ki correlations from past work.
Please let us know if isoNAM Ki you obtained from IC50 greatly differs from that using initial rate experiments (we have isoNAM Ki from initial rate experiments);
XG(11/18):
The isoNAM Ki for SIRT3 obtained from initial rate experiments is 4622 uM based on Mixed competitive inhibition model fitting.
RC: The numbers are quite different for initial rate and endpoint. In the literature you looked at, are initial rate experiments used?
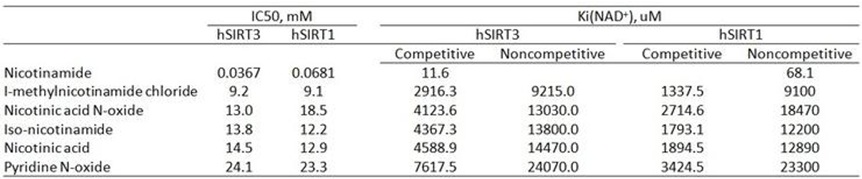
The Ki converted from IC50 value are listed in the table below.
The reference used for the calculation is listed below plus the website. Link is shown as following: http://botdb.abcc.ncifcrf.gov/toxin/kiConverter.jsp
- of expts with isoNAM will be planned after getting updated PLOS data analysis
PL:
*Prepare the SIRT3 data set with more quantitative information (e.g. by-residue RMSD);
*Run MM-GBSA calculations to obtain energetic information;
*Prepare MD simulation for SIRT3/Intermediate/NAM simulation from ternary structure by cleaving NAM and formation of new C-O bond;
PMC-AT Group Meeting 11112014 PLIN.pptx
-Comment on constrained loop sampling options in prime.
-Some papers claim loop conformational change after cleavage is important for next step of deacetylation and also for NAM release. Can we confirm this (at least former - see PNAS 2004 Sir2 xtalography paper) based on SIRT3 structures?
-Compare C pocket interactions of NAM in SIRT3:peptide:NAD+ to NAM in SIRT3:intermediate:NAM
-Multiple sequence alignment of sirtuin loop and C pockets
PL(11/17): I used 1YC2 (Sir2Af2) chain:B structure to identify the NAM binding pocket (C pocket), and carried out structural base sequence alignment using pdb structures for SIRT1, SIRT2, SIRT3, yHST2, SIRT6, Sir2TM, Sir2Af2.
The result is attached with the C pocket residues selected. The structure is highly conserved. alignment1.pdf
The multiple sequence alignment using HST1-4 of Yeast, and Sir2Af1, Sir2Af2, human sirtuins 1-7 etc, was performed using ClustalW2, and these residues are found to highly conserved. (the sections that involves these residues are presented below.)
RC: Does this include the loop? Is NPD_Thema Sir2Tm and NPD1,2_ARCFU Sir2Af1,2?
PL(12/3/14): Yes. The loop is included in the top, starting from the residues that I squared out.
Yes, NPD_Thema refers to Sir2Tm and NPD1,2_ARCFU correspond to Sir2Af1,2.
-As I noted in a recent email regarding the paper plans, we may be including some of our results on the computational prediction of ligand binding affinities and correlation with respect to experimental data. For this purpose, I would like to start by revisiting the existing C pocket binding affinity data for the series of C pocket binding ligands we studied starting with Eric's work. Please indicate where I can find the correlations with XG's experimental data and whether this included MM-GBSA,PBSA, and glidescore correlations. After reviewing these, I will provide further instructions on whether we need to continue this work with new binding affinity calculations and associated experiments for the purposes of the paper(s).
PL(11/24): I can not locate the Schrodinger's project file of EK's work on C pocket binding affinity calculation. Here is what I found from wiki "Docking Simulation" page.
RC: You will need to redo these dockings yourself if the files are missing and you haven't done them yourself - shouldn't take long. Also, you had compared MM-GBSA,PBSA, and glidescore rankings for another set of ligands in the past. We should calculate all three for the C pocket ligands below. Please schedule this. Next steps will be provided thereafter.
Thur., Apr. 18, 2013
Some of the results for SIRT3 using the newer PDB:4FVT crystal structure.
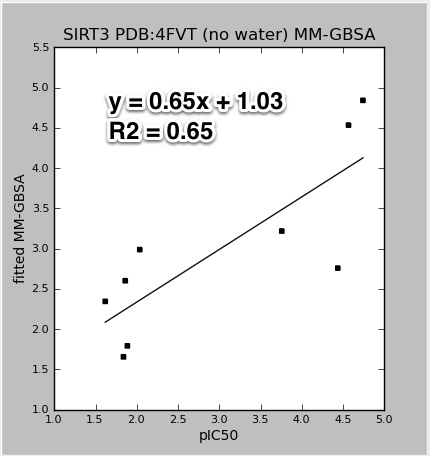 |
| 4FVT_MM-GBSA.vs.pIC50.jpeg |
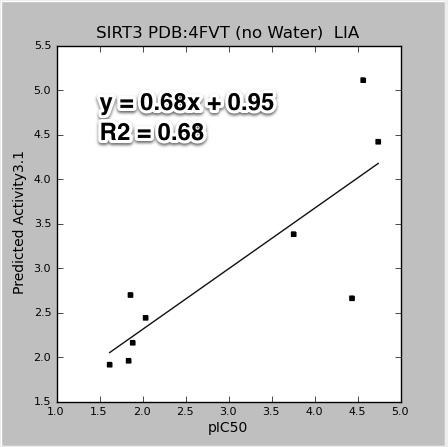 |
| 4FVT_LIA.pIC50.jpeg |
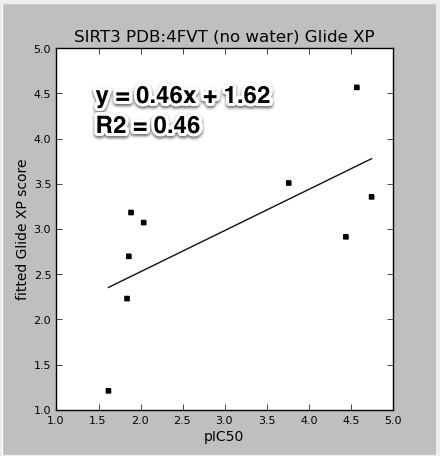 |
| 4FVT_GlideXP.vs.pIC50.jpeg |
The data points for the above figures are for all the SIRT3 inhibitors: AC93253, Salermide, EX-527, 1-methylnicotinamide, nicotinic acid N-oxide, iso-nicotinamide, nicotinic acid, pyridine N-oxide, and nicotinamide. The results would improve with the nicotinamide outlier (at pIC50 4.4) left out. Also, all the models do an impressive job accounting for the wide range of activities. Except for nicotinamide, all the smaller single ring molecules have much lower inhibition in the mM range. It is not clear to me if these good results would hold up with decoy molecules (molecules that are similar to potent inhibitors, but are known experimentally to not bind).
Also note that the Y-axis in the above plots are not the raw GlideXP, or MM-GBSA scores. The linear regression fits and scales those scores to correspond to the same range as the pIC50. Plots for the publication should not have this scaling. I will fix.
Monday, Feb. 18, 2013
Relationship between K_i and IC_50: linear, with few exceptions, depending on assumptions of kinetic model. The following paper shows detailed equations for the relationship between K_i and IC_50 for Michaelis-Mention kinetics for competitive, non-competitive and uncompetitive inhibitors. It also shows cases for very tightly bound inhibitors. In almost all cases, there is a linear relationship between K_i and IC_50.
- Cer, R.Z., Mudunuri, U., Stephens, R., and Lebeda, F.J. (2009). IC50-to-Ki: a web-based tool for converting IC_50 to K_i values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res 37, W441–W445. Link to paper.
Delta_G of binding for the inhibitors of SIRT3 from Xiangying's experiments.
Assuming Delta_G = RT ln K_i where R=1.99E-3 kcal/(K*mol) and T = 295K, and K_i is converted from uM to molar.
Note that there is no minus sign in the above equation, as binding and inhibition occur in opposite directions. If K_b for binding were used in the equation, there would be a minus sign.
| hSIRT3 inhibition |
Ki(NAD+), uM |
Delta_G (kcal/mol) |
(kcal/mol) |
(kcal/mol) |
||||
| Competitive |
Noncompetitive |
Competitive |
Noncompetitive |
GlideXP |
MM-GBSA |
|||
| Nicotinamide |
11.6 |
-6.66 |
-5.0 |
-31 |
||||
| 1-methylnicotinamide chloride |
2916.3 |
9215 |
-3.42 |
-2.75 |
-4.3 |
-40 |
||
| nicotinic acid N-oxide |
4123.6 |
13030 |
-3.22 |
-2.54 |
4.2 |
-15 |
||
| Iso-nicotinamide |
4367.3 |
13800 |
-3.18 |
-2.51 |
-4.3 |
-33 |
||
| Nicotinic acid |
4588.9 |
14470 |
-3.15 |
-2.48 |
-3.2 |
-22 |
||
| Pyridine N-oxide |
7617.5 |
24070 |
-2.86 |
-2.18 |
-2.2 |
-24 |
||
GlideXP scores are in the correct ball park of -2 to -5 kcal/molWhen comparing these Delta_G numbers to the simulation estimates of Delta_G with the below plots of experiment vs. docking for hSIRT3 inhibitors:
MM-GBSA scores are much lower (-15 to -40 kcal/mol) than absolute binding affinities. However, low numbers like these are reported in many other publications, such as the Cardozo paper.
Tuesday, Feb. 12, 2013
Experiment vs. Docking of hSIRT3 inhibitors (nicotinamide, 1-methyl nicotinamide chloride, nicotinic acid N-oxide, iso-nicotinamide, nicotinic acid, pyridine N-oxide) for both GlideXP and MM-GBSA. The results do not show a clear trend and do not show a better way to estimate the binding affinity through linear regression of docking binding affinity estimates to experimental binding affinity.
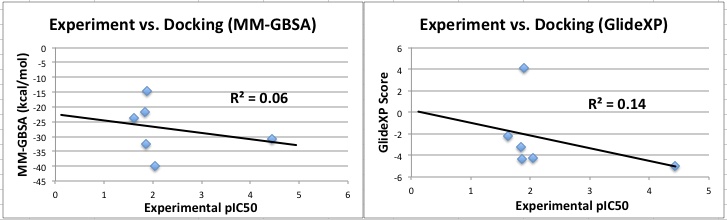 |
| Experiment.vs.docking.hSIRT3.jpeg |
Mon. Jan. 28, 2013
The following are results from docking ligands into SIRT3 (PDB: 3GLR). All of the below ligands docked to the C-pocket without constraints. However, some of the ligands had one or two higher ranked poses that were not in the C-pocket. The below table shows the highest ranked scores that docked in the C-pocket. The last two columns show the best ranked MM-GBSA scoring pose that is not in the C pocket. The other pocket for all cases is in between the B and C pockets (just above the C pocket). Those labeled N/A did not have any poses docked into other volumes besides the C pocket. In only one case (Nicotinic Acid) did the pose docked into the other pocket score higher with MM-GBSA than the C pocket pose. Of note, also, is the discrepancy between the GlideXP scores for Nicotinic Acid N-oxide.
| Pocket ----> |
C |
C |
Other |
Other |
| Ligand |
GlidXP |
MM-GBSA |
GlideXP |
MM-GBSA |
| Nicotinamide |
-5.00 |
-30.8 |
N/A |
N/A |
| iso-nicotinamide |
-4.33 |
-32.5 |
N/A |
N/A |
| 1-methylnicotinamide chloride |
-4.28 |
-40.0 |
-2.83 |
-21.1 |
| Pyridine N-Oxide |
-2.20 |
-23.8 |
N/A |
N/A |
| Nicotinic Acid |
-3.24 |
-21.8 |
-2.78 |
-28.3 |
| Nicotinic Acid N-oxide |
+4.15 |
-14.7 |
-3.30 |
-11.9 |
--Please provide estimated scheduling for the above tasks within the next couple of days; a table of tasks with estimated dates can be prepared. Priorities can be adjusted by RC after receiving proposed schedule--
(some tasks above were repeated from 10/11-1) below)
-Consider preparation of movies for visualization of MD sims
-Matlab license question of AS. Checking AS comments/questions and providing any required replies for next week's work.
schedule meeting to discuss next steps for MM-GBSA code dev after PL verifies that AS has run existing code and batch servers properly. as should also schedule next tasks for sysadmin (like secure ftp server administration
after some more of these done will schedule addition of gpu node to batch server
Consider capacity usage of GPU node for MD simulations and recommend whether a new gpu node should be purchased. If so, may involve AS in the planning/purchasing.
| 10/11 Group Meeting Presentation Reminders The following is not a complete list. Other information from experiments and simulations underway can be provided as well, as appropriate. Many of the computational reminders below are restatements of previously posted tasks. 1) Computational a) i- All pending info on the dataset for IFD should be provided at group meeting and also on wiki ii- All unanswered questions on tasks page should be answered on wiki before group meeting b) Provide the following at the group meeting: i- First provide the residue-by-residue RMSD for each residue in loop and also NAM between the SIRT3:peptide:NAD+ and SIRT3:intermediate:NAM xtal structures (see group meeting minutes from ~ 1 month ago) ii- It was indicated on tasks page that SIRT3:Intermediate:NAM MD simulations (starting from intermediate xtal structure) were previously run. Use ptraj in Amber to provide times series of RMSD with respect to the initial structure as well as the products of the NAM cleavage reaction (separately for both NAM and flexible loop). iii- From the results of MD simulation starting from intermediate superimposed in the SIRT3:peptide:NAD+ xtal structure (currently running), provide time series plots of RMSD with respect to NAM and loop in intermediate complex, as well as analogous plots with respect to starting structure; also provide the time series plot of the associated energies. iv- Also provide residue-by-residue B factors and NAM B factors from these simulations c) The following simulation should be set up and ready for discussion at meeting (it was originally requested in place of b-iii above): i-MD simulations starting after NAM cleavage reaction initialized by energy minimization after bond cleavage, retaining NAM in the complex. Compare NAM pose to that prepared by docking to this same structure, prior to starting the simulation. Related: ii- compare C pocket interactions of NAM in SIRT3:peptide:NAD+ to NAM in SIRT3:intermediate:NAM iii- provide single point energy for products of the NAM cleavage reaction (from c-i starting structure, after minimization) iv- provide ensemble average energies for intermediate:NAM complex (once available from c-i after equilibration) (iii,iv should be provided in a table. Both MM-PBSA and MM-GBSA values can be provided) d) Multiple sequence alignment of all sirtuin C pockets with listing of residues interacting with NAM and how (latter from structural analysis) 2) Experimental (can start preparing these for group meeting and finish after meeting) a) Literature data on sirtuin experimental kinetic characterization i- Please provide a list of all sirtuins with their IC50s for NAM ii- Also maximum inhibition by NAM for all these sirtuins iii- All known info on rate constants of all these sirtuins in the same table including which step is rate limiting if known iv- Also make a table with all known mutants and activity effects, including which mutants were studied computationally v- A 2007 paper by Denu measuring the NAM cleavage rate constant by rapid quench methods and radioactivity may have been missed. Please check. Will need to later update the ppt on experimental kinetic characterization accordingly. b) Protein purification i- Alok should start planning expression and purification of SIRT3 (and possibly SIRT2 if time permits; to be discussed) to be done alongside b-lactamase purifications AU: I need plasmid containing SIRT3/2 gene with His Tag. It can be purchased from commercial sources. I told Dr. Guan about Origene and they have OFR's in mammalian expression system. We can ask them (or any other vendor for quote etc) ii- Alok may start to review the SDS-PAGE gels provided by Enzo and consider, with Guan, possible sources of the impurities (point for discussion) AU: I looked at SDS PAGE, lookes like Enzo is not interested in much purer protein, they are more happy with specific activity. Yes, this can be discussed in group meeting. Minutes of Group meeting (10/17/2014) AU: Previous Work: Summarize Zhen's work, what she had done or what kind of data she had and if she made any protocol, look in to Q&A. (I will look again in her files and make a summary of it and post on wiki) Current Work:AU-Group Meeting 10-17-2014.ppt Computational modeling was done with Cephalothin. Growth Assay in liquid culture verses on plates specially when presenting in a paper.(Both methods are valid in manuscript, MIC table can be made in both cases and if reviewer asks about figure etc, I can go to plate for "representative data"). RC: It would be a good idea within the next couple of weeks to do at least one comparison between plate and liquid to make sure the results are comparable. AU: Thanks, I will do it along with some mutants, so that I compare both WT and few mutants. Why Method 1 (for growth assay) worked with 10 microL inoculum not with 5 microL? (10 microL cells may have enough to survive initial antibiotic concentrations). Sequence all clones which are positive for IPTG inducible proteins. Prioritizing mutants characterization: Decision has to be made either from computational side or from biochemical side. If computational side has some high scoring mutants based on algorithm then I can use them. From Biochemical side, I can categorize mutants based on their MIC values such as 1: equal to WT, 2: >WT, 3: <WT and then feedback to refine the model if necessary. Adding 1 or 2 more antibiotics in the screening. See file containing structure of antibiotics Zhen mentioned in her spread sheet. Structures.docx (waiting for Dr. Chakrabarti's suggestions; two thing to consider, 1: different b-lactam class and widely used in literature so that we have comparative analysis. Protein Purification: How much time it will take to purify a protein? It takes 3 days to get protein ready to use. I can purify 5 proteins at a time if resources are available. XG: PMC-AT Group Meeting_10.17.2014.pptx 1) Prism Fitting for PLOS ONE data at different NAM concentration. RC: For future experimental design, it will be useful to do further fittings with subsets of PLOS ONE data, including fewer [NAD]+ and more [NAM] and analysis of effects on accuracy of parameter estimates including alpha. 2) Double reciprocal plot_0NAM, 50uM DHP 1c (0uM NAM, 50uM NAM) RC: Please include these in the ppt below when ready. 3) Plan for another set of experiments of [NAM]=100uM at 0uM and 560uM DHP 1c 4) Plan endpoint experiments for DHP 1b and 1b derivative on SIRT1 snd SIRT3. RC: Please provide this somewhat in advance of finishing all other planned experiments so there is sufficient time to consider implications for next set of experiments. PL: PMC-AT Group Meeting 10172014 PLIN.pptx Prioritize the work including 1) SBIR proposal, 2) Sirtuin modeling projects, and 3) Arabinda's task assignment and management. (Details will be provided on Ping's Task List.) RC: Comments on ppt and results to date on 2). Some comments refer to previous group meeting minutes below. Sirtuin structure prediction validation: - Dataset for systematic structure prediction methods development and testing should be provided prior to starting validation. This dataset can be provided on the paper 2 page as discussed. It should include a list of available sirtuin structures (esp SIRT3, but can also include other sirtuins if the SIRT3 dataset is too small), including ligand (drug) complexes wherein loop conformational shifts occur and for selected pairs of such structures, indicate the relevant RMSDs of active site side chains and relevant secondary structural motifs including the flexible loop. These can be used in testing. (Please indicate B factors from xtalography where relevant.) The superposition of structures shown e.g. in the latest group meeting ppt are difficult to interpret visually. See tasks page for more details on how to organize the dataset. - For each such structure pair, one structure will be used as initial to predict second structure (this includes pairs where the structures are the same and one seeks to predict in the native environment). In each case where structure prediction accuracy is poor we are interested in determining if this was due to scoring function inaccuracy, inadequate sampling, or structural flexibility (energetic degeneracy). - Eventually, for paper 2, it will be important to do (customized) IFD protocol testing on structurally characterized ligand complexes such as Ex-527 after above are completed to assess false positive rate. E.g., we should be able to show that preferred Ex-527 binding mode is not buried below the C pocket. For such scoring the IFD scoring function may need to be modified as discussed. Sirtuin structure prediction IFD scoring functions: - After the dataset above is prepared and while some initial simulations are running, further information on the parameter estimation and testing protocols used to develop and validate induced fit methods in the literature should be summarized. In particular, when did Schrodinger test its advanced IFD protocol and with what dataset. - A note: We have been planning to develop customized scoring functions for sirtuin structure prediction. In the present context, this can be used to improve structure prediction accuracy on the above testing dataset (need to first assess dataset size and break into training and testing subsamples). - For later scaling of IFD calculations, note that we have PLOP (older gen prime) source code available and it is used in python protein design script. This can be run on an arbitrary number of nodes. Hence we can do extensive sampling of protein degrees of freedom in our protocols (moreso than ligand degrees of freedom for now) and may later modify IFD to use PLOP not prime as needed. AS may be involved in this. Conformational changes upon NAM cleavage (please indicate when you plan to schedule these tasks): - Analysis of past MD simulation data on NAM in C pocket of intermediate complex should be provided as indicated below after our previous group meeting. Please provide analysis of B factors for loop and NAM and a comparison to NAD+ (ternary) complex structure (see below for more details). As noted below MD simulations of the conformational change between these complexes can follow after these results are reviewed and discussed as needed. These are relevant to paper 2. - Differences in flexible loop between intermediate and ternary complex not yet clear. Is the comparison provided on 4BVG and 3GLS between intermediate and apo, from two different studies? How many intermediate structures are available and from which studies (this may be answered in context of dataset above)? Does the loop conformation differ so substantially in all structures if there are more than one? Please summarize the differences. Please also indicate whether a loop prediction is being run starting from ternary complex structure with NAM cleaved. - Summaries of MD results, as they become available, should also be organized systematically in a section of wiki page for 2nd paper. - MD simulation results can also be used to prepare movies suitable for viewing by AS and other group members for purpose of visualizing MD results. - Please present the results from the loop predictions underway at time of group meeting, as they become available. - As indicated below results of MD simulations of loops (when they are done) should be compared to results of highest ranked results from ab initio loop prediction to assess adequacy of loop sampling of the long loop (see below for more details). Minutes of Group meeting (9/29/2014) AU:AU-Group Meeting 9-29-2014.ppt Discussion regarding growth assay, induce protein expression in liquid media before plating on LB-plates containing IPTG Repeat transformation in BL21 cells. Look in to Zhen's data if she has done screening for mutants.: [Unable to locate assay/MIC data for mutants] RC: They were not sequenced, so the data is not very useful, but there was probably some MIC data in tables provided with Zhen's report that Chaoran obtained. You can ask Sherry about this report. It is not a priority right now in case you cannot find it. AU: I found only WT data Wildtype MIC 6 substrates.xlsx, there are 3-4 excel sheet but it doesn't say what mutant etc (not named properly e.g. Colony plate summary sheet v2.xlsx). Why amount of protein expressed differ from clone to clone. XG:PMC-AT Group Meeting_09.29.2014.pptx 1) Finish [NAD+]=750uM, 1500uM, and 3000 uM with 50 uM DHP 1c. 2) Pick higher [NAM], like 100uM for next round experiments. PL: As a general principle, we need to start using multiple available structures for validation of computational structure prediction methods (starting w validation of known mechanism-based inhibitors). The dataset is of considerable size and as many structures as possible could be used for validation. Esp with IFD, there is a concern regarding false positives. In general, such problems may be broken down into issues with a) sampling; b) accuracy of the energy function. Both will be assessed in the context of sirtuin structure prediction. Current focus is on rank ordering of structures; later binding affinity datasets may be used. 1) Review MD simulation study in PLOS ONE_ loop motions after NAM docked? -check what kinds of loop motion we have sampled in our MD simulations of NAM in C pocket (summary statistics); what were starting structures? what were timescales over which significant loop motions were observed? Visualization of trajectories (lower priority) -Summarize the structural differences (NAM and loop) between NAD+ and intermediate from literature and xtals -Related (lower priority), check Wolberger's "NAM flipping" conjecture vis-a-vis this analysis - compare flipped conformation to that we observe and indicate what side chains are involved in stabilizing the flipped conformation 2) Energetic issues, induced fit calculation -When ranking protein structures obtained by rotamer sampling or ligand poses only, same degrees of freedom are sampled for each ranked structure. We need to revise IFD protocol to be able to properly incorporate protein reorganization energy in scores used to rank order binding modes. We could, e.g., start by modifying script to keep track of exactly what residues are being sampled and then sample the same ones for different ligand poses. Same number of docking/protein structure prediction iterations should be used for each ranked binding mode. MD could then be used on structures prepared in this way for binding affinity estimation (receptor preparation also needed for reporting absolute binding affinities). See also (3) below regarding the energy function. -Before moving on to new IFD calculations, we need to test such a protocol on pairs of sirtuin structures wherein protein conformation changes occur (to be selected). E.g., could test ability to predict Ex-527 conformation in co-product complex starting from a different receptor structure with important differences in protein conformation. In any such test, include scoring of the crystallographic structure to determine whether a test failed because of inadequate sampling or an energy function error. -After further discussion, relevant programming tasks will be assigned to AS 3) formula of IFD score MM-GBSA score may be used in place of docking score. 4) Revisit Ex-527 PNAS paper: loop open/close conformation 5) Thinking about possible MD simulations to exam (A)EX527:coproduct issue; (B) start from intermediate after the cleavage of NAM. Former is for purposes of computational methods validation; latter is a step of the sirtuin catalytic cycle that has not been studied computationally. Results of 1,4) will be useful in advance. -For (A), check if loop prediction can confirm that for Ex-527/coproduct complex, the drug stabilizes the closed conformation of the loop. Check if ab initio loop prediction ranks highly those loop conformations that appear most frequently in MD simulations. -For (B), set up the MD simulation for at least as long as the timescale in 1). Further discussion will be possible after an update is provided on the results from 1). Provide summary statistics on loop and NAM conformations, comparing to intermediate xtal, and possibly visualize trajectory. Prepare initial structure by breaking/making the bonds manually, setting appropriate charges, and optimizing geometry. Confirm the length of the flexible loop. Assuming the intermediate complex structure differs significantly from that of NAD+ complex, try to predict the former from the latter using loop prediction, or at least whether the xtal loop structure is one of highly ranked ab initio structures. Energy minimize the xtal loop conformations and compare them to the ranked loop energies to determine effect of sampling efficiency vs energy function accuracy. Again compare the results to those from MD. If successful, MD or ab initio loop sampling may be useful in customized IFD protocols (to be subsequently automated) using a limited number of pre-generated loop conformations to reduce backbone sampling space. Constraints may also be applied later to allow limited sampling in this regard. Even if not successful, similar customized sampling algorithms may be developed. (Possible issues with experimental prep protocols for xtals and loop disorder will be discussed later.) Some of this analysis may be relevant to paper 2. Other tasks related to continued outlining of subsections of this paper will be posted later. PMC-AT Group Meeting 09292014 PLIN.pptx Prepare Arabinda's tasks and/or software development wiki pages on PMC-AT wiki. Note that there were many notes regarding prior software dev on the academic wiki (webdesign-ror). AS will be invited to that as well, and may copy immediately relevant info to this wiki. PL can post the tasks for AS and AS management listed by RC last week to one of these pages. After PL has decided whether to test the modified IFD script with license requests only and has updated on this, RC can review cluster NFS/NIS configuration and then RC/PL can expand upon/finalize AS tasks vis-a-vis above points as needed. Meeting schedule with AS will be decided at this time as well. Minutes of Group meeting (9/12/2014) AU: Discussion about activity assay in cell lysate or purified enzyme? Will be decided later on. IPTG optimization in liquid culture then decide how much to use on plates. update sequencing data once available. Since I am using DH5alpha, then we will know sequence information (mutants) before we do growth/activity assay. It will be precise but labor intensive. AU-Group Meeting 9-12-2014.ppt AU-Sequencing Primers.doc XG: 1) Purchase DHP-1b and test the activity; 2) Different time point assay (one [NAD+], and one [modulator]); 3) Update on wiki the NAM concentration calculations; 4) Plan on experiments: a) DHP-1c activity test at 1 uM on SIRT1 (EC150_SIRT1 = 1uM); b) Dosage dependent assay to test how high [DHP 1c] can go for SIRT3 (EC150_SIRT3=50uM); c) 2nd manuscript preparation; 5) Will calculate NAM concentration to decide what [NAM] need to be used for relative inhibition assay; 6) CRO quote for enzyme-peptide:compound binding affinity measurement. Enzyme cost vs. time for expression and purification of enzyme in house; 7) Future work: more kinetic study on certain molecule in the presence of NAM. Will decide after the preliminary experiments done. PMC-AT Group Meeting_09.12.2014.pptx PL: 1) Upload the files into the Dropbox; PMC-AT Group Meeting 09122014 PLIN.pptx RC (9-14): To include in list of induced fit computations for this week: -SIRT3:NAD+:peptide - DHP (4FVT) -Compare DHP binding affinities for modes w, w/o intermediate or NAD+ (e.g., 4BVG induced fit calculation reported was in presence of intermediate; compare DHP binding affinities to those in absence of intermediate, where DHP may extend into A pocket). -Please indicate: did you run any induced fit calculations with inhibitors, such as Ex-527? If so, did you identify the buried binding mode? If not, please check whether induced fit identifies a buried binding mode for Ex-527, e.g. for SIRT3:intermediate Minutes of Group meeting (9/5/2014) Experimental- AUAU-Group Meeting 9-5-2014.ppt
(1) Modifications on 2nd manuscript
PMC-AT Group Meeting_09.05.2014.pptx 2nd paper_xg_9.4.14.docx RC: At the next group meeting, please provide an estimate of the length of the part of the paper up to the end of the basic kinetic model section (without the derepression model sections, as previously discussed) so we can assess whether the paper should be split at that point. Please also post the transient kinetics data that you recorded during the endpoint assays for both no DHP and DHP so we can compare the initial rates if possible. XG(9/8): The current assay provide the activity at 1 hr incubation instead of quenching reaction at different time points. To obtain more information of the relationship between [NAM] and activation, the different time points queching experiment can be done. Have you been working on the continuous assay? XG(9/8): Not for last week. Will continue upon complishment of DHP .testing Computational-PL * C binding pocket of SIRT2 and SIRT3 have more than 90% high similarity base on sequence and structure alignment result. * In the structure of SIRT2:DHPs (no ADP) partial loop region is missing (3 residues) * Effect of peptide binding does not do much in terms of DHP binding * With APO enzyme the structure is more open, DHPs do not stay deep. * Induced fit docking, does it sample that loop? Does the software decide which residues to sample? what the box size? For our docking can you add the missing loop first then dock? * Manually sampling vs. common induced fit sampling protocol PMC-AT Group Meeting 09042014 PLIN.pptx RC: Please post the group meeting slides when ready. Minutes of Group meeting (8/22/2014) Experiment Part *Consider to use TCan for long term solution *Calculate dosage of radioactive reagent *Find out the procedure for license application *Cost for continuous assay vs. Fluor-de-Lys kit *Provide more details on how to cooperate Dr Raj. commons into the draft of 2nd paper , no need to be complete but show progress. PMC-AT Group Meeting_08.22.2014.pptx PMC-AT Group meeting 082214 PLIN.pptx Simulation Part *Screening of EX-527 like small molecules_1300 commercial available compounds were docked by Glide and re-ranked based on MMGBSA calculation. Top 20 molecules are presented. PMC: How long did it take to complete the job? PL: ~ 2 weeks. PMC: Are the results from only SIRT3? How about SIRT1? PL, RC: The SIRT1 structure has not been fully resolved yet. Computationally there is a limitation. PMC: Can we test the potential candidate in the lab experimentally?XG: Yes. RC: Are they from same receptor? Is this only for inhibitor? PL: Yes. SIRT3:Co-product is the receptor. Yes, it is for inhibitor. RC: Recalled using core hopping Schrodinger, 40,000 compounds were generated, MMGBSA calculation had been completed. PL needs to summarize the results at some point. PMC: Are there any overlapping between the libraries generated by MOE and core hopping Schrodinger? PL: No. For example, there is always “–Cl” in those from Schrodinger, but not MOE at last in different position. RC: Ideally the scores for the top 200 molecules in each list along with the scoring function used will be provided (see previously posted wiki comments). Also the molecules most resembling Ex-527 may be highlighted. RC: Any plan for screening more molecules? PL: (1) for the leads generated by MOE, we can re-dock and calculate MMGBSA for the purpose of comparison. (2) Patent search for new lead and then similarity search for commercial available compounds, dock, MMGBSA calculation… *Next step is to do the patent search PMC, RC: Looking for commercial available source for patent search, like patent search engines. Set a phone conference with patent lawyer for free consultations. PMC, RC: New hiring in India may help. Lead to 3D open source; simplify the workflow; write script… *DHP compounds RC: PL needs to summarize from the MMGBSA, Glide scores PL: Inhibitor binds stronger in the absence of peptide because of more room structurally. RC: which job (MMGBSA calculation or Glide docking) is more time consuming? PL: MMGBSA RC: Try induced fit on different receptors to find more poses PL: Induced fit is more time consuming. It takes for 2 days. RC: Can we say that the peptide has bigger effect on SIRT2 than that on SIRT3 based on simulation results? RC: Whether there is difference at active site for SIRT2 and SIRT3 which directs to different binding mode? RC: Develop a correlation between MMGBSA scores and IC50 data. Ex527? DHP 1a-d? In another word, can be predict IC50 based on MMGBSA score? RC: PL should setup -Structural visualization? Sharing? How? RC (8-22): Some of the points above now need to be incorporated into the list of the tasks for the following week for each person which should be posted to the tasks pages. A couple of additional questions/points to address: a) Exptl: we need to discuss the NAM concentration during the endpoint assays in JMC paper. Please remind me at the appropriate time. b) Computational: the original lead lists were scored according to different scoring functions (e.g. HTVS). The top 200 or so compounds were then reranked. What type of info can you provide on the extent to which the rankings changed? If significant changes occurred, it is clearly desirable to rerank a larger number of the compounds from the lists, especially given the fact that they used very different approximations in the virtual screening. c) Computational: please indicate whether there could be any speed advantages accrued by using free software for MM-GBSA calculations, given the fact that these are the bottleneck and we have only one Schrodinger license. d) We may consider looking at the commercially available leads we generated from our similarity searches and check which ones have binding modes most similar to the DHP inhibitors, and then including all these molecules in a series for which we compare IC50/binding affinity correlations. RC: Ping, please share the folder with maestro files for visualization as discussed. Also, please incorporate points from the above notes into your task list for the week and post. The following is also of particularly high priority and relevant additional files as generated should be added to the shared visualization folder: e) provide more info on the buried C pocket binding mode for SIRT2 that was shown at the group meeting. Indicate the extent of overlap between this binding mode and the A pocket. Do a structure alignment with SIRT3. Using SIRT3:intermediate structures where appropriate, show the clashes that may occur between this binding mode and the intermediate. Is this binding mode stable for SIRT2 structures prepared from both apo and SIRT2:peptide complexes? More comments/tasks pertaining to more extensive sampling of protein and ligand degrees of freedom and more accurate binding affinity calculations given the existence of multiple stable binding modes will be posted subsequently. DHP IC50's for SIRT2 have been provided in JMC paper at 500 uM NAD+. We will assess consistency between computationally predicted binding modes / affinities and experimental data shortly. Group meeting 6-3 RC overview and objectives: XG experimental section PMC-AT Group meeting_06.03.2014_experimental section.pptx RC: Ping, please post the computational part here as well. Guan, when you get a chance, please check some of the leads from the computational part against the Ex-527 patent to see if they are covered by the patent. groupmeeting_June2014_PLIN.pptx Schrodinger Princeton User Meeting - May 15, 2014 (Ping Lin attended.) May 15th, 2014 Schrödinger Regional User Meeting, Princeton, NJ The Schrödinger Princeton User Meeting will be held at the **Princeton Marriott at Forresta****l**on May 15th, 2014. As in past years, the regional user meetings offer a great opportunity to see current research from your colleagues and to learn about the latest developments from Schrödinger. Agenda 9:00 am Characterization of ligand-bound conformations with QM-MM methods: Application to prolyl hydroxylase enzymes Alejandro Crespo, Merck |
|||||||
| 9:40 am |
How to treat waters in virtual screening? A case study on the adenosine A2a receptor Bart Lenselink, Leiden University |
||||||
| 10:20 am |
Coffee Break |
||||||
| 10:40 am |
The role of explicit solvent in the molecular recognition of a non catalytic-site HIV Sarah Boyce, Schrödinger |
||||||
| 11:20 am |
Vibrational circular dichroism as a tool to assign the absolute stereochemistry of drug molecules Ed Sherer, Merck |
||||||
| 12:00 pm |
LiveDesign: A new paradigm in scientific enterprise software for drug discovery Matt Wessel, Schrödinger |
||||||
| 12:15 pm |
Protein-Ligand Database (PLDB): An authoritative repository of structural data and protein-ligand interactions Braxton Robbason, Schrödinger |
||||||
| 12:30 pm |
Lunch |
||||||
| 1:30 pm |
KEYNOTE: FEP-guided lead optimization Bill Jorgensen, Yale University |
||||||
| 2:30 pm |
Free energy perturbation (FEP): Finally ready for large-scale application in drug discovery Woody Sherman, Schrödinger |
||||||
Note 1: The free energy perturbation implemented in Schrodinger using a combination of several techniques to make the workflow easily accessible to non-experts and obtain results comparable to the MCPRO method used by Jorgensen's group.
1) REST (Replica Exchange with Solute Tempering, for enhanced sampling within 5ns/window)
2) OPLS2 (improved force field on torsional energy and charge method - now using CM1A-BCC method, and automatic parameter generation tool - FFBuilder to set up new parameters)
3) FEP mapper (for automatic design the perturbation paths for congentic series of compounds)
4) Desmod GPU implementation
5) Cycle Closure (for error estimation)
WORKSHOP – Bryn Mawr College - March 13, 2013 (Ping Lin, Xiangying Guan, Chaoran Jin attended.)
Park Science Building, 101 N. Merion Ave., Bryn Mawr, PA 19010 MAP
====================================================
9:00-12:00 Designing Inhibitors with MOE Structure-Based Drug Design ToolsVisualization / Pharmacophores / Docking / Combinatorial synthesis
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
The course covers the application of in silico structure based drug design (SBDD) tools for the rational design of Tarceva-based EGFR kinase inhibitors. Starting with raw PDB protein-ligand 3D structures, all the steps required to initiate and advance a SBDD study are covered: preparing PDB structures for modeling, binding pocket visualization, protein-ligand contact analysis and the use of SAR for in situ modeling (modifying and optimizing ligands in the binding pocket) to design new compounds. Advanced topics such as pharmacophore query generation, protein-ligand docking, protein alignments for binding site comparison and in situ combinatorial synthesis will also be covered.
====================================================
13:30-17:00 Small Molecule Virtual Screening and AnalysisMolecular alignments / Pharmacophore discovery / Reaction based screening / Docking
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
The course covers applications for fragment-based drug discovery, lead optimization and library design. Equally applicable to ligand-based and structure-based projects, examples include the replacement of a scaffold, fragment linking, fragment derivatization and growth as well as the application of medicinal chemistry transformations for lead optimization. The course also includes the application of organic synthesis based virtual combinatorial library enumeration for synthesizing compound libraries and evaluating potential candidates during lead optimization, which is further enhanced with pharmacophore-based filtering to maintain key interactions with a receptor.